Schematic representation of CoBDock blind docking workflow. The docking... | Download Scientific Diagram

Fully Blind Docking at the Atomic Level for Protein-Peptide Complex Structure Prediction. | Semantic Scholar
Protein-Ligand Blind Docking Using QuickVina-W With Inter-Process Spatio-Temporal Integration | Scientific Reports

Blind docking to the whole cavity of the hERG ion channel (A). Focused... | Download Scientific Diagram

Prediction of ligand binding sites using improved blind docking method with a Machine Learning-Based scoring function - ScienceDirect
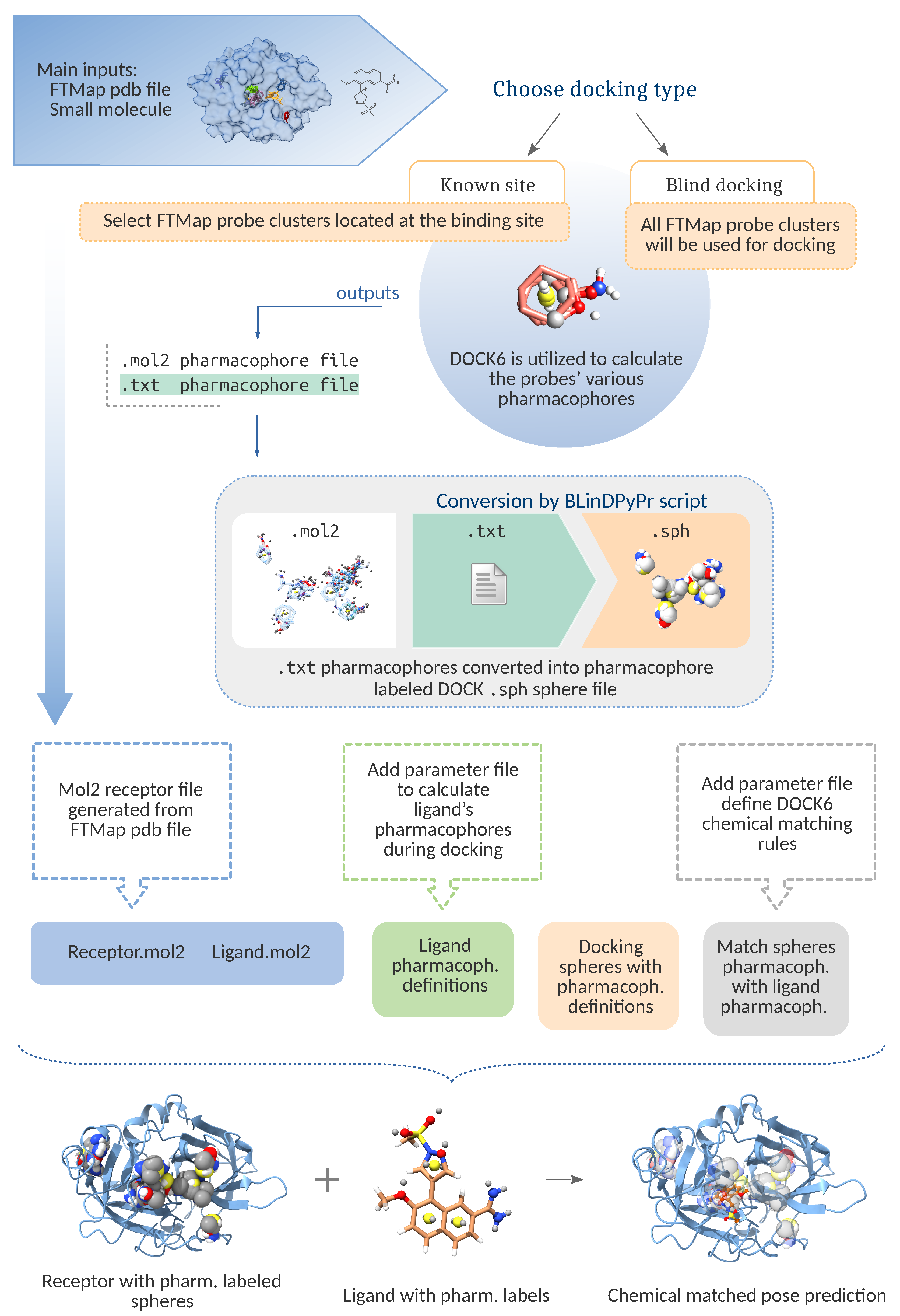
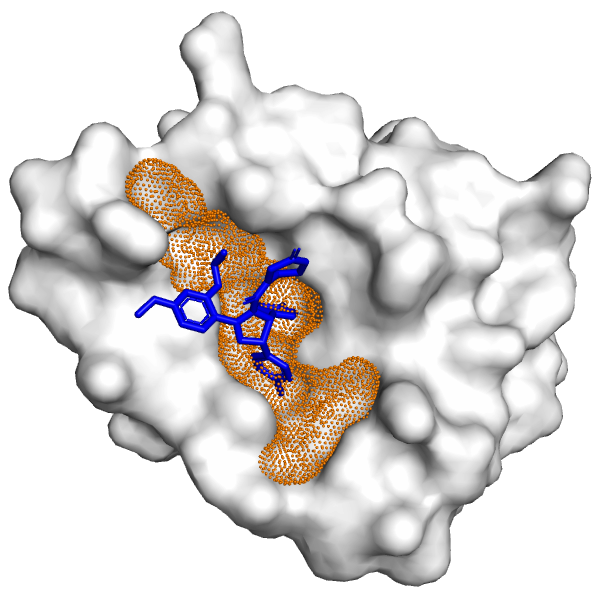
Molecules | Free Full-Text | Improving Blind Docking in DOCK6 through an Automated Preliminary Fragment Probing Strategy
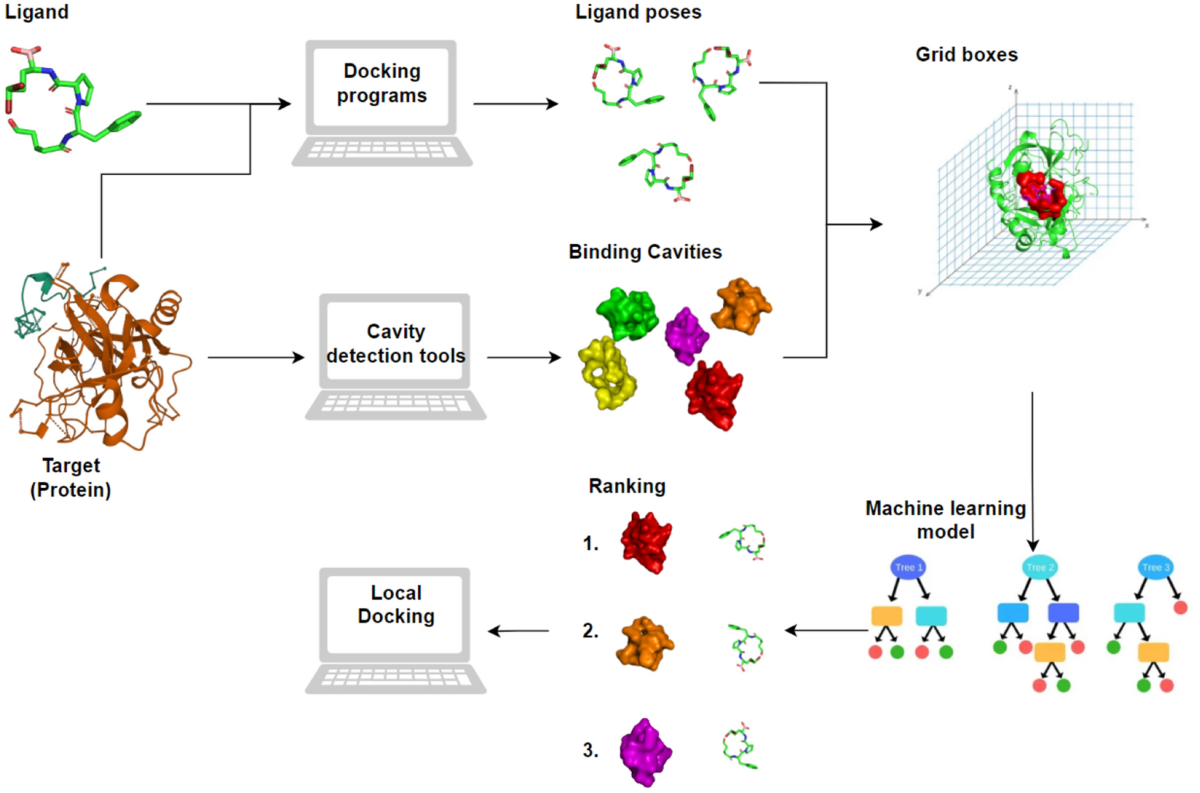
Cobdock: an accurate and practical machine learning-based consensus blind docking method | Journal of Cheminformatics | Full Text

Fully Blind Docking at the Atomic Level for Protein-Peptide Complex Structure Prediction - ScienceDirect

Blind docking at exhaustiveness 32 (a) Different binding conformations... | Download Scientific Diagram
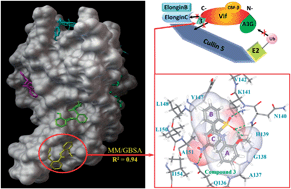
Exploring the binding mode of HIV-1 Vif inhibitors by blind docking, molecular dynamics and MM/GBSA - RSC Advances (RSC Publishing)
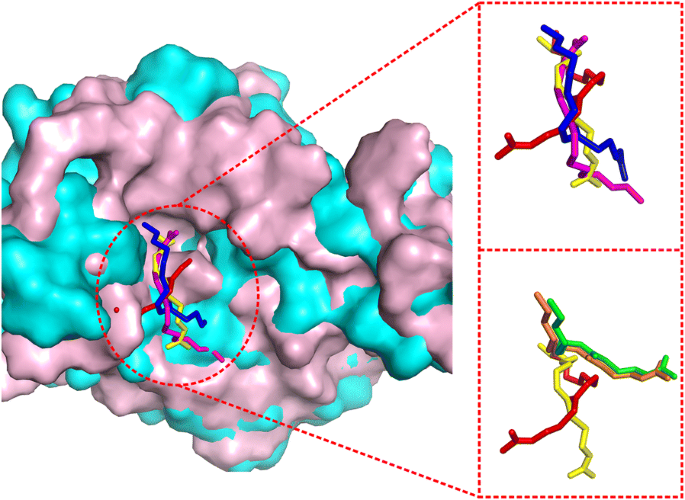
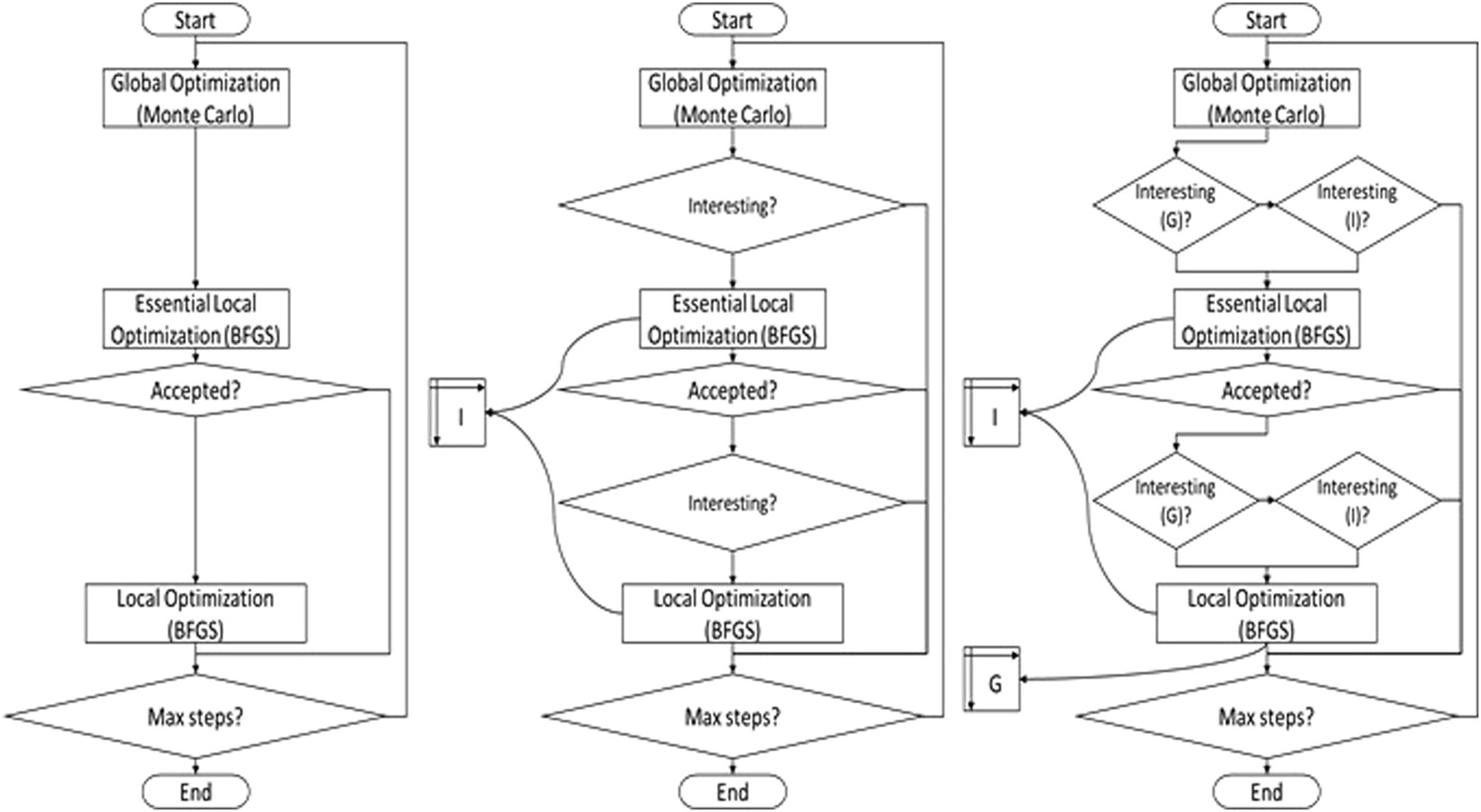
EDock: blind protein–ligand docking by replica-exchange monte carlo simulation | Journal of Cheminformatics | Full Text

Fully Blind Docking at the Atomic Level for Protein-Peptide Complex Structure Prediction. | Semantic Scholar

Blind docking on 5K7O: (A) Conformations distributed throughout protein... | Download Scientific Diagram